Structure-based drug discovery of GPCRs
GPCRs are a family of approximately 800 membrane proteins in the human genome. They mediate the majority of cellular responses, and are the targets of nearly half of today’s pharmaceuticals. Recent advances in structural biology have lead to GPCR structures in different functional states. I’m interested in using these high-resolution structures to discover GPCR ligands that confer new signaling properties. With our collaborators, I’m seeking novel ligands that target both the orthosteric and the allosteric sites of GPCRs.
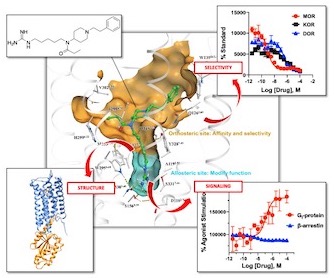
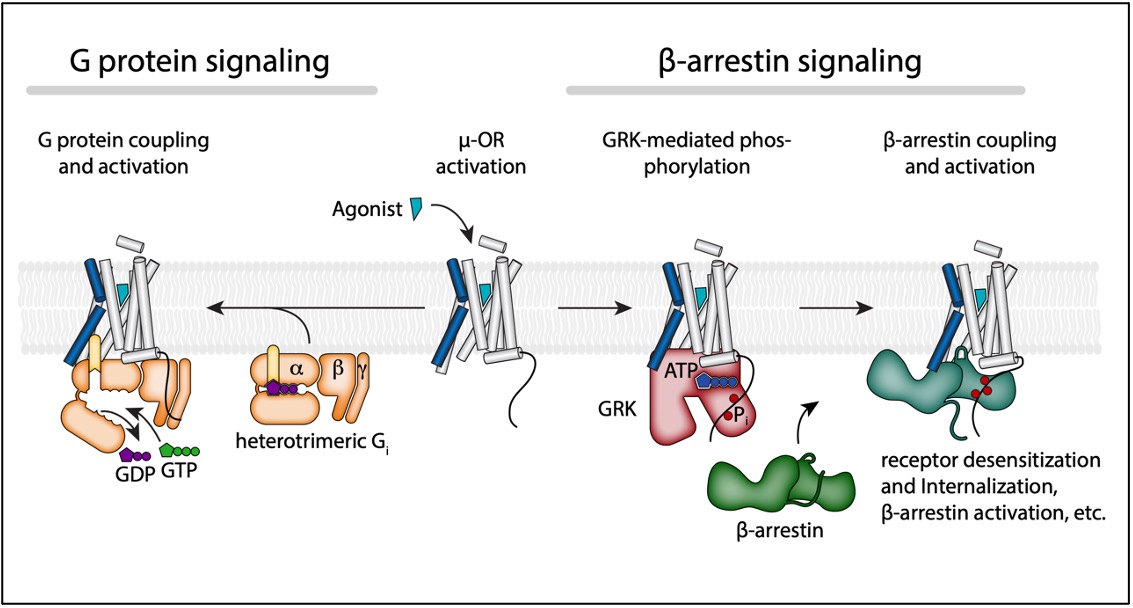
Biased signaling of opioid receptors
Opioid receptors are the major drug target for opioid analgesics. Activation of opioid receptors initiates signaling through a G protein pathway by activating heterotrimeric Gi, as well as through arrestin pathway by G-protein-coupled receptor kinase (GRK)-mediated phosphorylation and β-arrestin coupling. Biased agonism describes the preferential activation of one pathway over the other. In vivo studies suggested that G protein biased agonism for opioid receptors might provide therapeutic advantages. However, the molecular mechanism by which G protein biased agonists selectively avoid arrestin pathway is not well understood. I’m interested in how different parameters affect phosphorylation and β-arrestin coupling by integrating biochemistry and structural biology. The proposed research will make progress towards the understanding of biased agonism, and ultimately provide a foundation for the design of functionally selective analgesics with fewer adverse effects.
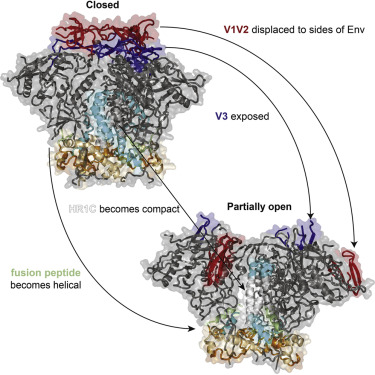
Conformational changes of HIV-1 Env trimer during viral fusion
I’ve solved structures of Env in prefusion and receptor-bound states, described a detailed structural biology model of Env-mediated membrane fusion. Structural study of HIV-1 Env in receptor-bound state was known to be challenging due to its dynamic property. To overcome this issue, I used a combination of bNAbs to stabilize the complex of Env and CD4 receptor. This strategy enabled us to obtain atomic resolution structures of CD4 receptor-bound Env. These structures answered some fundamental questions, such as how HIV-1 Env undergoes conformational changes during receptor and coreceptor engagement and how these conformational changes could lead to viral-host cell membrane fusion.
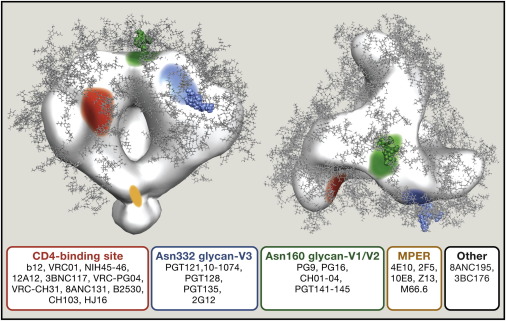
Broadly neutralizing antibodies against HIV-1 virus
Broadly neutralizing antibodies (bNAbs) neutralize HIV-1 virus by targeting Env glycoprotein on viral surface. Previous studies have suggested that administration of bNAbs after HIV infection can lead to viral clearance in animal models. In order to understand the neutralizing mechanism of these bNAbs, structural studies are essential. My early work focused on characterizing bNAbs isolated from HIV-1 patients. As a graduate student in Bjorkman lab, I have solved multiple structures of patients’ bNAbs in complex with Env glycoprotein. These results not only demonstrated the neutralizing mechanism of bNAbs, but also revealed novel epitopes on HIV-1 Env. The knowledge of how bNAbs recognize epitopes on HIV-1 Env can guide antibody engineering as well as vaccine design.